In recent years there has been an explosion of parallel algorithms for solving bioinformatics problems, namely phylogenetic reconstruction and sequence alignment. These algorithms follow the growth of new hardware solutions like Field-Programmable Gate Arrays (integrated circuits capable of performing simple instructions in parallel), Cell microprocessors (like the one inside Playstation 3), Graphics Processing Units (nvidia and ATI powerful graphic cards) and massively parallel cluster architectures (like the IBM BlueGene). There is now an article describing a parallelized Needleman–Wunsch alignment algorithm for the the Tile64 RISC processor.
The Tile64 card is composed of 64 core processors, with each core running its own Linux OS and standard programs, and communicating using the Tilera API. The Tile64 is a System on Chip (SyC), that therefore can be plugged into a PCI slot and be used independently from the CPU. On the other hand it can handle only integer number instructions, which limits its usability for numerical computations.
The Needleman–Wunsch algorithm is used for global sequence alignment. That is, for given two sequences it tries to maximize the score by including as few insertions as possible in each one of the sequences. It is closely related to the Smith-Waterman algorithm for local alignment, which tries to find the longest subsequence with positive score – where the score function is almost the same as for Needleman–Wunsch.
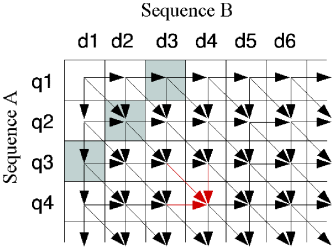
Both algorithms are a dynamic programming method where a matrix is built with the scores for all possible pairwise combinations (the solution is found by backtrack after the matrix is complete). After initialization of the matrix (first row and first column) the score of a cell can be calculated by looking at its immediate top and left neighbor cells, represented by the arrows in the figure below. For example the score of cell q4d4 depends only on q4d3, q3d3 and q3d4.
In the article they use an implementation of the FastLSA algorithm, a parallel version of Needleman–Wunsch where instead of storing the whole matrix it stores one row/column combination per block, since depending on the sequence length the memory requirements for the whole matrix can become prohibitive. In other words it stores the score values only for a grid of rows and columns (e.g. at every ten sites). In [1] they claim that this implementation is therefore well suited for very long sequences, which cannot be handled for instance by the “needle” application of the EMBOSS package or the CUDA implementation of the SmithWaterman algorithm [2].
The parallelism is achieved if we notice that the cells belonging to the same anti-diagonal (one such anti-diagonal represented in gray) can be calculated independently. Thus distinct cores can calculate the score of these cells at the same time with the so-called wavefront parallelism. Their solution achieved gains of 20 times over similar programs – even though their SyC implementation is in C and the other CPU implementations are in Java.
references:[1] Galvez, S., Diaz, D., Hernandez, P., Esteban, F., Caballero, J., & Dorado, G. (2010). Next-generation bioinformatics: using many-core processor architecture to develop a web service for sequence alignment Bioinformatics, 26 (5), 683-686 DOI: 10.1093/bioinformatics/btq017
[2] Manavski, S., & Valle, G. (2008). CUDA compatible GPU cards as efficient hardware accelerators for Smith-Waterman sequence alignment BMC Bioinformatics, 9 (Suppl 2) DOI: 10.1186/1471-2105-9-S2-S10


No comments:
Post a Comment
Use the space below to ask, inform and criticize -- if you are not very happy please read the rules for commenting.
Please, do not include unrelated, commercial sites not even in your signature.